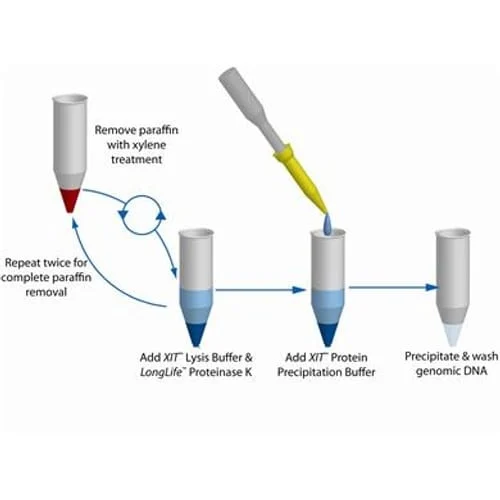
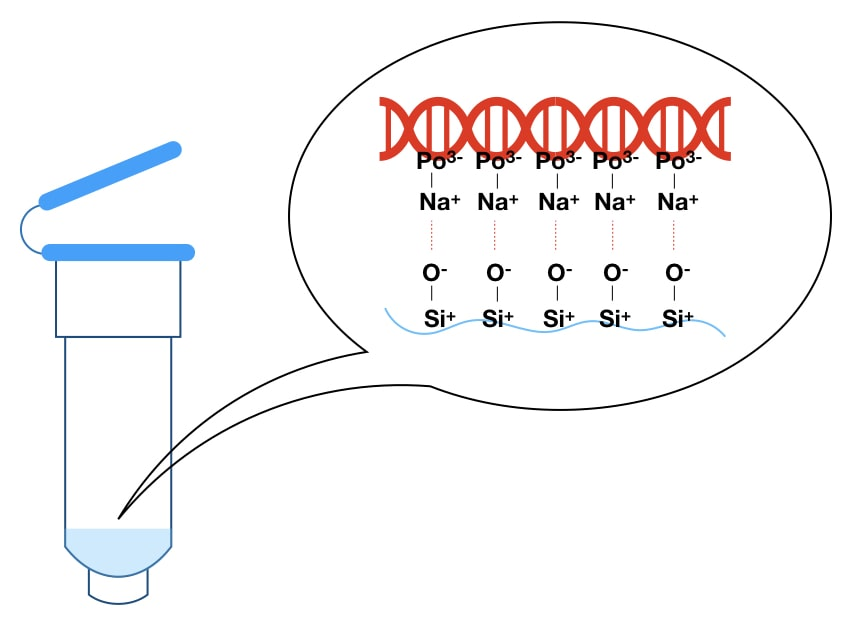
DNA extraction is a fundamental procedure in molecular biology, providing the foundation for a wide range of genetic analyses. Whether the goal is to sequence a genome, identify genetic mutations, clone a gene, or analyze a forensic sample, the quality of the extracted DNA plays a crucial role in the success of these downstream applications. The process of DNA extraction involves isolating DNA from cells or tissues, separating it from proteins, lipids, and other cellular components, and purifying it for further use. Although the basic principles of DNA extraction are well-established, achieving high yields of pure DNA can be challenging, especially when dealing with different sample types or large-scale extractions. This comprehensive guide explores the key techniques, common challenges, and best practices for mastering DNA extraction in the laboratory.
Fundamental Techniques of DNA Extraction
DNA extraction can be carried out using various techniques, each with its own advantages and limitations. The choice of method depends on several factors, including the type of sample, the required DNA purity and yield, and the intended downstream application. Below are some of the most commonly used DNA extraction methods:
- Organic Extraction (Phenol-Chloroform Method)
The organic extraction method, often referred to as the phenol-chloroform method, is one of the oldest and most reliable techniques for isolating DNA. It involves the use of organic solvents, typically phenol and chloroform, to separate DNA from proteins and other cellular debris. During the process, cells are lysed using a detergent, such as SDS (sodium dodecyl sulfate), which disrupts the cell membrane and releases the DNA into the solution. The mixture is then treated with phenol and chloroform, which form two phases: an organic phase (containing proteins and lipids) and an aqueous phase (containing nucleic acids). After centrifugation, the DNA is recovered from the aqueous phase and precipitated using ethanol or isopropanol.
Advantages:- High-quality DNA: Organic extraction yields high-quality, intact DNA, which is suitable for sensitive applications like PCR, sequencing, and cloning.
- Effective for complex samples: This method is particularly effective for extracting DNA from samples with high levels of contaminants, such as tissues rich in proteins or lipids.
- Time-consuming: The process involves multiple steps, including phase separation and precipitation, making it more time-consuming than other methods.
- Hazardous chemicals: The use of toxic organic solvents like phenol and chloroform requires careful handling and proper disposal.
- Silica-Based Extraction (Column or Membrane Method)
- Cost: Commercial silica-based kits can be expensive, especially for large-scale or high-throughput applications.
- Yield: While the purity is high, the yield may be lower compared to other methods, particularly for samples with low DNA content.
Limitations:
- Simplicity and speed: Silica-based methods are straightforward and can be completed quickly, making them ideal for routine DNA extraction.
- High purity: The DNA obtained is usually free of contaminants like proteins and RNA, making it suitable for most molecular biology applications.
Silica-based extraction is a popular method that uses silica columns or membranes to selectively bind DNA while impurities are washed away. The process typically involves lysing the cells, binding the DNA to the silica under high salt conditions, washing away contaminants with ethanol-based buffers, and eluting the purified DNA in a low-salt buffer or water.
Advantages:
- Magnetic Bead-Based Extraction
- Equipment requirement: This method requires a magnetic separator or automated system, which may not be available in all laboratories.
- Cost: Similar to silica-based methods, the cost of magnetic bead-based kits can be high, especially for large-scale extractions.
Limitations:
- Automation-friendly: Magnetic bead-based methods are compatible with automated liquid handling systems, enabling high-throughput DNA extraction with minimal manual intervention.
- Reduced contamination risk: The use of magnetic beads reduces the risk of cross-contamination, as the beads can be easily separated from the liquid using a magnet.
Magnetic bead-based extraction is a modern technique that utilizes paramagnetic beads coated with DNA-binding ligands. The process involves lysing the cells, binding the DNA to the magnetic beads, washing away contaminants, and then eluting the DNA from the beads. This method is particularly suited for automated and high-throughput extraction.
Advantages:
- Chelex 100 Method
The Chelex 100 method is a simple and rapid technique primarily used for DNA extraction from small and challenging samples, such as forensic samples or buccal swabs. Chelex 100 is a chelating resin that binds metal ions, thereby protecting the DNA from degradation during the extraction process.
Advantages:- Speed: The Chelex 100 method is fast, typically requiring less than an hour from start to finish.
- Minimal equipment: This method requires minimal equipment, making it accessible for laboratories with limited resources.
- Lower purity: The DNA obtained may contain impurities and inhibitors, making it less suitable for some downstream applications.
- Limited sample types: This method is generally used for specific sample types and may not be suitable for all DNA extraction needs.
Challenges in DNA Extraction
While DNA extraction is a routine procedure, it is not without challenges. The following are some of the most common issues encountered during DNA extraction and strategies for overcoming them:
- Sample Quality and Integrity
The quality and integrity of the starting material are crucial factors that influence the success of DNA extraction. Degraded or poorly preserved samples can lead to low yields of fragmented DNA, which may not be suitable for certain applications. To ensure optimal sample quality:- Proper sample collection: Use appropriate techniques and tools for collecting samples, and avoid exposing the samples to extreme temperatures, UV light, or other environmental factors that could cause DNA degradation.
- Preservation and storage: Store samples at the correct temperature and in suitable conditions (e.g., freezing at -20°C or -80°C) to preserve DNA integrity until extraction.
- Inhibitors and Contaminants
DNA samples often contain inhibitors, such as proteins, lipids, polysaccharides, and other contaminants that can interfere with the extraction process or downstream applications. These inhibitors can co-purify with DNA, leading to poor yields or compromised results. To mitigate the impact of inhibitors:- Lysis buffer optimization: Use lysis buffers that are specifically formulated to break down the cellular components of your sample type, reducing the presence of inhibitors.
- Additional purification steps: Incorporate additional purification steps, such as proteinase K treatment or RNase treatment, to remove specific contaminants.
- Use of appropriate extraction methods: Choose an extraction method that effectively separates DNA from inhibitors. For example, organic extraction is effective for samples with high protein content, while magnetic bead-based methods are suitable for automated systems with contamination control.
- Yield vs. Purity Trade-Off
A common challenge in DNA extraction is balancing yield and purity. High-yield extractions may result in co-purification of contaminants, while methods that prioritize purity may sacrifice DNA quantity. To optimize both yield and purity:- Optimize protocol parameters: Adjust parameters such as incubation time, temperature, and the volume of reagents to maximize both yield and purity.
- Evaluate multiple methods: Test different extraction methods on your specific sample type to determine which one provides the best balance of yield and purity.
- Quality control: Regularly assess the quality of extracted DNA using spectrophotometric analysis (e.g., A260/A280 ratio) and ensure that the DNA is suitable for your intended application.
Best Practices for Optimal DNA Extraction
Adhering to best practices during DNA extraction can significantly improve the quality and reliability of your results. Here are some essential practices to follow:
- Standardizing Protocols
Consistency is key to reliable DNA extraction. Standardizing protocols across all samples ensures reproducibility and minimizes variability. This includes following the same procedures for sample collection, storage, and extraction. Make sure to:- Develop and document SOPs (Standard Operating Procedures): Clearly outline each step of the extraction process and ensure that all lab personnel follow these protocols consistently.
- Regular training: Provide regular training for lab personnel to ensure they are familiar with the protocols and understand the importance of consistency.
- Using Fresh Reagents
The quality of reagents can significantly impact DNA extraction efficiency. Always use fresh, high-grade reagents, and regularly calibrate equipment to ensure optimal performance. Some tips include:- Store reagents properly: Store reagents according to the manufacturer's instructions to maintain their effectiveness.
- Avoid contamination: Use sterile techniques and aliquot reagents to avoid cross-contamination.
- Implementing Quality Control Measures
Incorporate quality control steps to monitor the success of DNA extraction and identify any issues before proceeding to more complex analyses. Essential quality control measures include:- Measuring DNA concentration and purity: Use spectrophotometry or fluorometry to assess the concentration and purity of extracted DNA. The A260/A280 ratio is commonly used to evaluate purity, with a ratio of 1.8-2.0 indicating pure DNA.
- Gel electrophoresis: Run extracted DNA on an agarose gel to assess its integrity and size distribution. Intact, high-molecular-weight DNA should appear as a distinct band with minimal smearing.
- Optimizing Lysis and Vortex Mixing
The lysis step is critical for releasing DNA from cells, and the efficiency of this step can greatly influence the yield and quality of the extracted DNA. To optimize lysis:
- Ensure complete lysis: Use appropriate lysis buffers and conditions (e.g., temperature, incubation time) to ensure complete cell disruption.

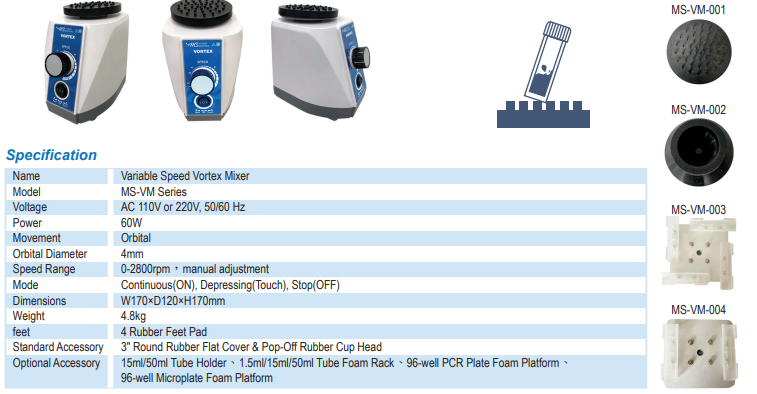
- Vortex mixing: Incorporate vortex mixing during the lysis step to ensure thorough mixing of the sample with the lysis buffer. Vortex mixing generates a controlled spiral flow in the liquid, promoting uniform cell lysis and efficient DNA release.
- The Major Science Vortex Mixer series is an efficient and compact instrument for any routine mixing applications needed in the field of life sciences, chemistry, medicine, bioengineering, analytical and research laboratories. This product includes the eccentric rotating sturcture to generate vortex flow in the liquid in test tubes centrifuge tubes, microplate and other containers and offers fully mixing the solution. Choose from two modes of operation : continuous mode provide variable speed can allows low rpm start-up for gentle mixing or high speed mixing for vigorous vortexing of samples, touch mode which activates mixing when depressing the platform. The Major Science Vortex Mixer series is stable and reliable with reliable durability and also has a variety of accessories to choose from to adapt to different experimental needs
DNA extraction is a cornerstone of molecular biology, serving as the foundation for countless applications in genetics, forensics, biotechnology, and medicine. By mastering the various techniques, understanding the challenges, and adhering to best practices, you can achieve consistent, high-quality DNA extractions that enable accurate and reliable downstream analyses. Whether you are working with challenging samples, optimizing protocols for high-throughput workflows, or ensuring the highest standards of purity and yield, the principles outlined in this guide will help you navigate the complexities of DNA extraction and achieve success in your molecular biology endeavors.